Figure: The progression of antimicrobial resistance, from exposure to global transmission.
Published: November 2025 | Author: Elias Hossain
Antimicrobial resistance (AMR) has become a defining biomedical challenge of our century, a silent pandemic that grows stronger with every misused antibiotic. While traditional genomic models can classify resistant strains, they often lack interpretability, the ability to explain the reasoning behind predictions. My current research focuses on bridging this gap using Large Language Models (LLMs) fine-tuned for biomedical data, making AMR prediction both accurate and explainable.
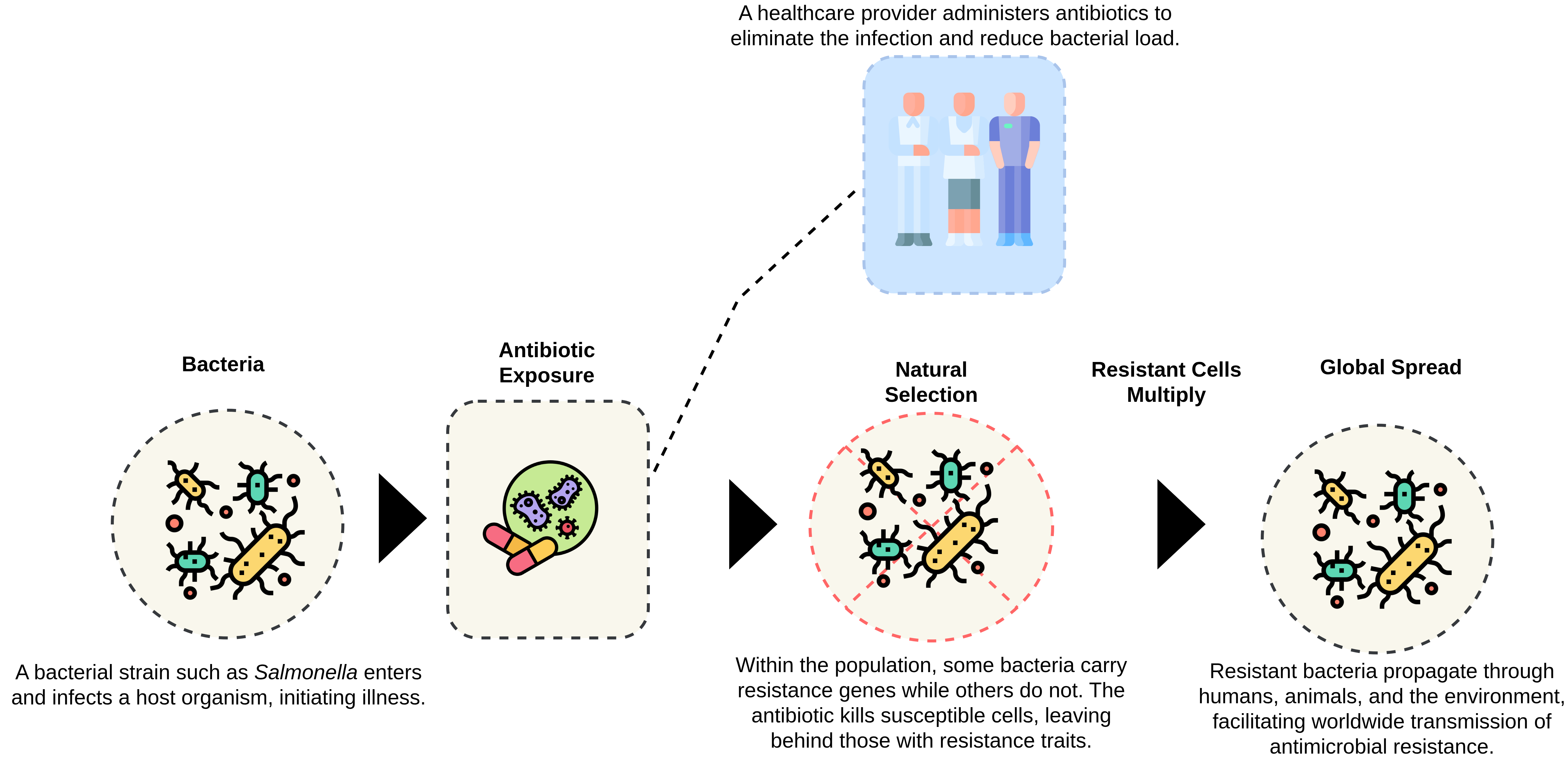
During my early experiments, I noticed that resistance is not just a sequence-level phenomenon. It is ecological, behavioral, and evolutionary. To visualize this, I designed a simplified model of bacterial adaptation that connects exposure, mutation, and spread.

Figure: The progression of antimicrobial resistance, from exposure to global transmission.
The diagram shows how exposure to antibiotics triggers selective pressure. Susceptible bacteria die, while resistant ones survive and replicate. Over generations, these survivors dominate the population, eventually spreading resistance genes across species and even environments. Understanding this progression inspired me to model resistance as an evolving language, one that LLMs could decode through genomic and semantic cues.
In my framework, I integrate biomedical LLMs such as BioBERT and PubMedGPT with molecular embeddings to infer resistance pathways. The model does not stop at predicting, it generates human-readable rationales like:
These insights bridge machine predictions and biological interpretation, helping researchers trace resistance at both the molecular and evolutionary scales.
Transparent AI models in microbiology can guide clinicians toward better treatment strategies, reduce diagnostic uncertainty, and identify potential drug targets earlier. More importantly, interpretability builds trust, a fundamental requirement for AI integration in healthcare.
From: Elias Hossain
Ph.D. Student, University of Central Florida (UCF)